This is a source code and data release for the ACM Siggraph 2013 paper Practical SVBRDF Capture In The Frequency Domain, by Aittala, Weyrich and Lehtinen.
Most of the code is written in Matlab and requires some toolboxes to run (image processing, preferably also parallel toolbox for performance). The capture tool is C++ code and requires the Canon EDSDK. The code is research code, and hence unfortunately is not particularly readable, flexible or efficient. We hope that you will find it useful nevertheless.
Copyright (c) 2013-2015 Miika Aittala, Jaakko Lehtinen, Tim Weyrich, Aalto University, University College London. This code and data is released under the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International license (http://creativecommons.org/licenses/by-nc-sa/4.0/).
Source code (Matlab/C++)
Dataset: Mix (~700 MB)
Dataset: Pynchon (~700 MB)
Dataset: Crumpled (~700 MB)
Dataset: Eco (~700 MB)
Dataset: Tile (~700 MB)
Dataset: Bluebook (~700 MB)
Raw TIFF images for Lindt dataset (3.5 GB)
The source code archive contains the following directories:
capture_toolC++ source for the software that displays the on-screen patterns and drives the camera. Requires Canon EDSDK.
optimizerMATLAB source for building the dataset from the raw images, specifying calibration, and computing the SVBRDF.
dataPlaceholder for datasets.
outputPlaceholder for output data.
The actual optimization is performed by optimizer.m. It shows the current iterate (starting at a low resolution) and gradually improves it. Finally it outputs a Matlab data file sols.mat and the resulting images as 16-bit TIFFs in the specified directory. The displayed images are: diffuse albedo, specular albedo, kurtosis, normals, glossiness, unused (shows upsampling progress), and normals in another visualization. Notice that the glossiness is "inverse" (dark is shiny), as it corresponds to our sigma parameter.
For performance, you should launch a few parallel jobs using matlabpool (if you have it.) The optimizer outputs quite a bit of messy progress data into the console. Thanks to the low-resolution initial stage, you should be able to see rather quickly whether the solution is going to be something reasonable.
matlabpool 6
% Always include the final \ and make sure that the directory exists!
sols = optimizer(Data, 'Z:\whatever_your_path\mix\');
The solution will be in sols, which is a 1024 * 1024 * 10 array. The 10 channels are (for historical reasons): diffuse R, specular R, diffuse GB, specular GB, glossiness, normal x, normal y and kurtosis.
The tool that displays the patterns on the monitor and drives the camera is in the capture_tool/ directory. It is a Visual Studio 2010 project. It also requires the Canon EDSDK for camera control.
Again, this is a crude tool used for research and experimentation. A proper product-like implementation could be significantly more friendly and automated.
We have used a Canon EOS 5D Mk II camera. The temporal syncing between shutter and patterns has been chosen experimentally and may not match the times needed for some other camera model. The important thing is that the shutter must be open by the time the pattern starts showing, and must close after it has finished; preferably with some safety buffer at each end.
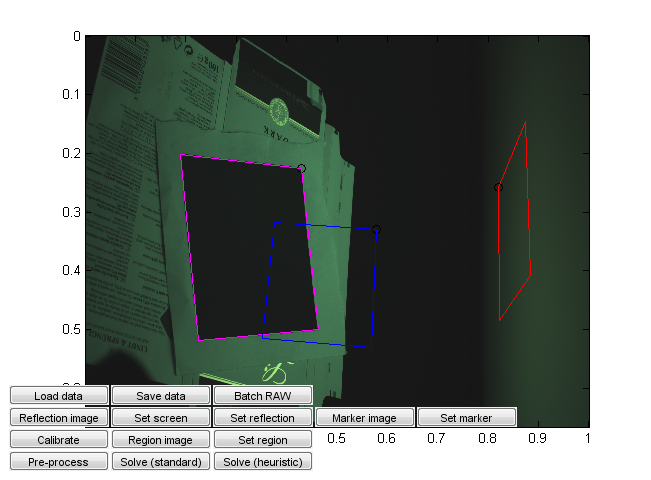
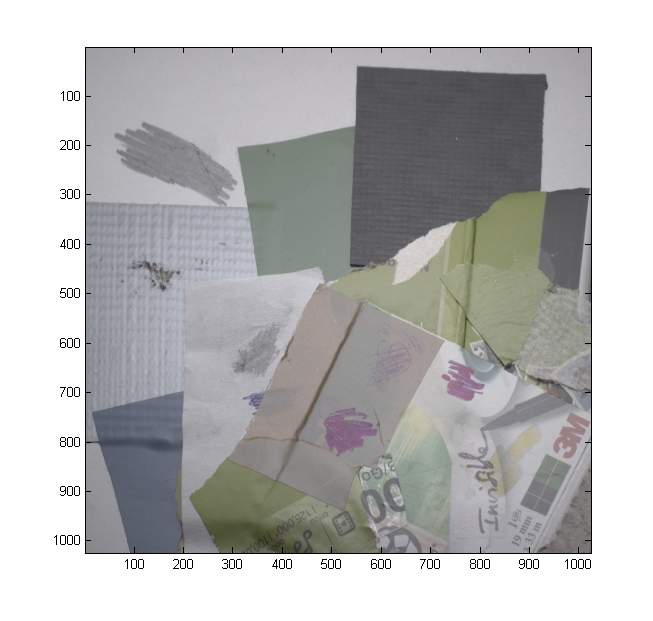
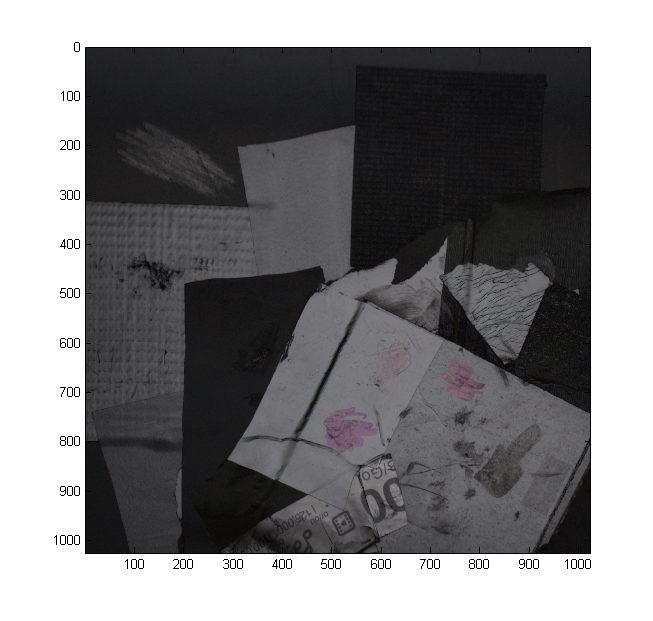
We have included a dataset (Lindt) that contains an entire output of a capture session as TIFF files converted from Canon RAW files by dcraw with maximally linear settings. Have a look at the photos (especially the first ones) to get an idea of what they should look like.
The overall steps for capturing are:
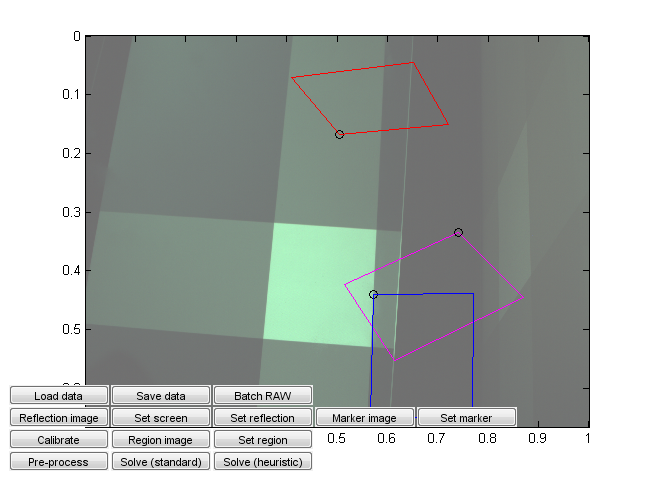
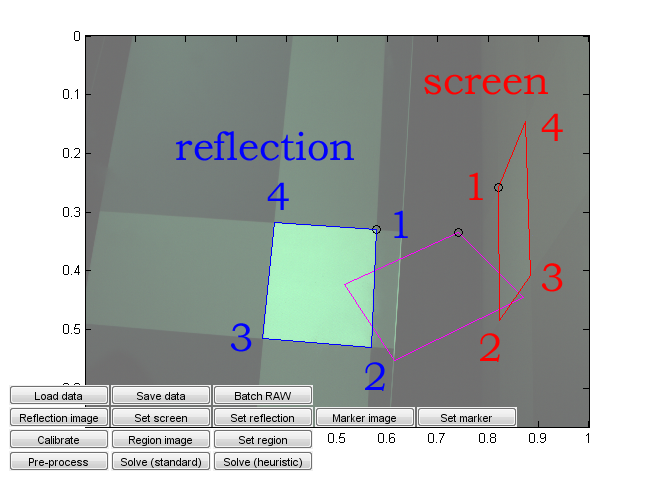
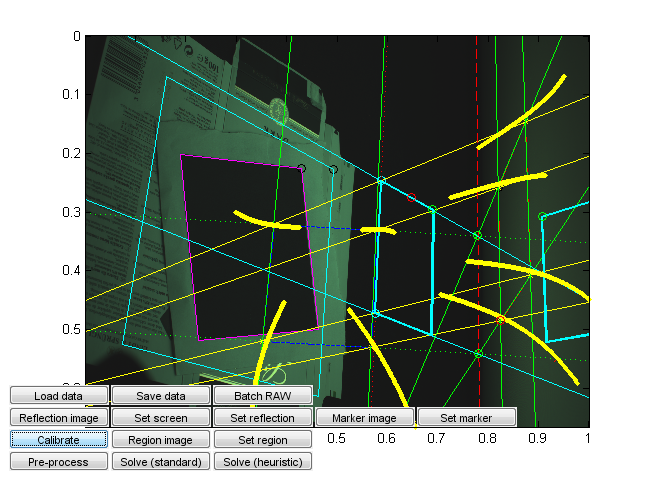
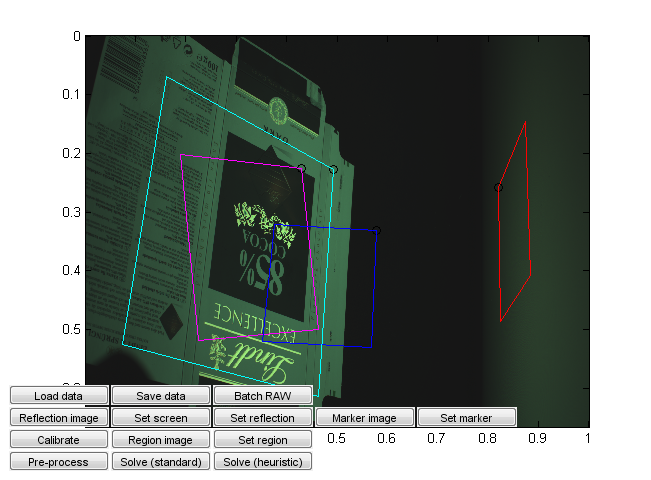
Run calibration_ui.m. This is a calibration and solver launch tool.
This, too, is a crude research tool. Note in particular that it is currently hardcoded assuming a 16:9 capture monitor aspect ratio and a given monitor emission distribution. Change the values from the code if needed. (You can probably recycle the monitor calibration values if they happen to work for you, but the aspect ratio should correspond to that used in the capture.)
The steps are:





The data packages each contain a single .mat file, which contains a Matlab struct. Let us use the Mix dataset as an example. Load it up:
load('path_to_data\data_mix.mat')
The data will be placed into a struct called Data. To see its contents, simply type Data. The struct contains the image data itself, and all relevant calibration information. For historical reasons, names and conventions do not always match to those in the paper.
Let's first familiarize ourselves with the near-raw data. The instructions for running the actual optimizer are in the end of this section.
To see the DC component (zeroth frequency image, i.e. simply illuminated by the plain window function), type
imagec(4*Data.DC);
(the factor 4 is just to make it a bit brighter; imagec is our simple color image viewing function which does gamma correction and is a bit less picky than Matlab's image)
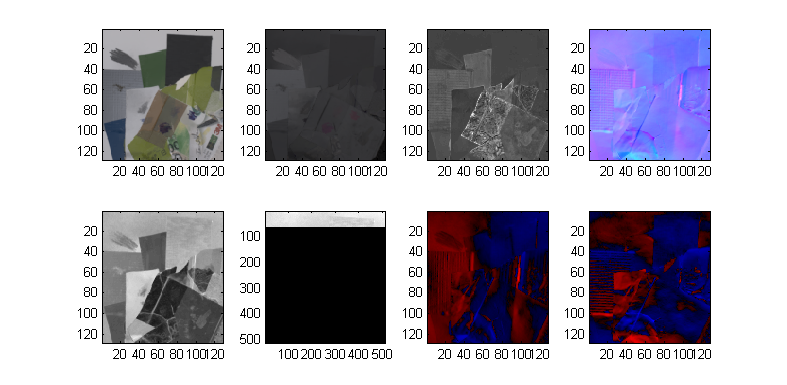
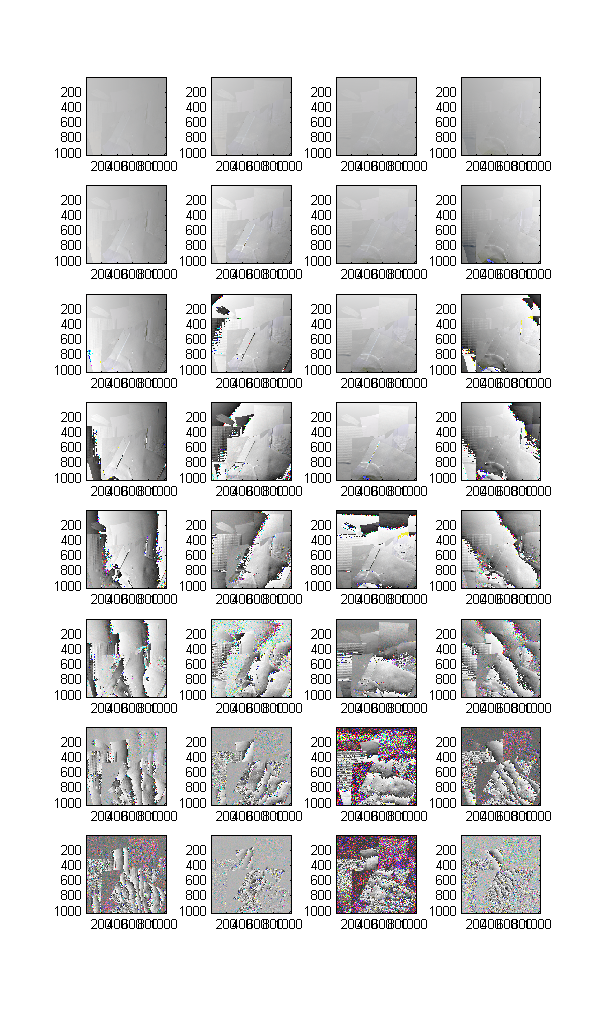
The actual frequency pattern images are stored as complex numbers in Data.Z, which is a 1024 px * 1024 px * 3 colors * 8 frequencies * 4 orientations array. The actual frequencies themselves are listed in Data.freqs. Let's see all the data images at once.
Here are the magnitudes, in a 8*4 array corresponding to orientation and frequency. Notice the effect of increasing frequency: the diffuse component fades away rather quickly.
% Magnitudes of all data
clf;
subplot(8,4,1);
for o = 1:4
for f = 1:8
subplot(8,4,4*(f-1)+o);
imagec(4*abs(Data.Z(:,:,:,f,o)));
end
end

Here's a representative example in higher resolution:
imagec(4*abs(Data.Z(:,:,:,6,3)));
Let's have a similar look at the phase. Clearly the phase seems to contain strong clues about the normals, but it is still distorted especially at low frequencies:
% Phases of all data
clf;
subplot(8, 4,1);
for o = 1:4
for f = 1:8
subplot(8,4,4*(f-1)+o);
imagec(angle(Data.Z(:,:,:,f,o))/2/pi+0.5);
end
end
imagec(angle(Data.Z(:,:,:,6,1))/2/pi+0.5);
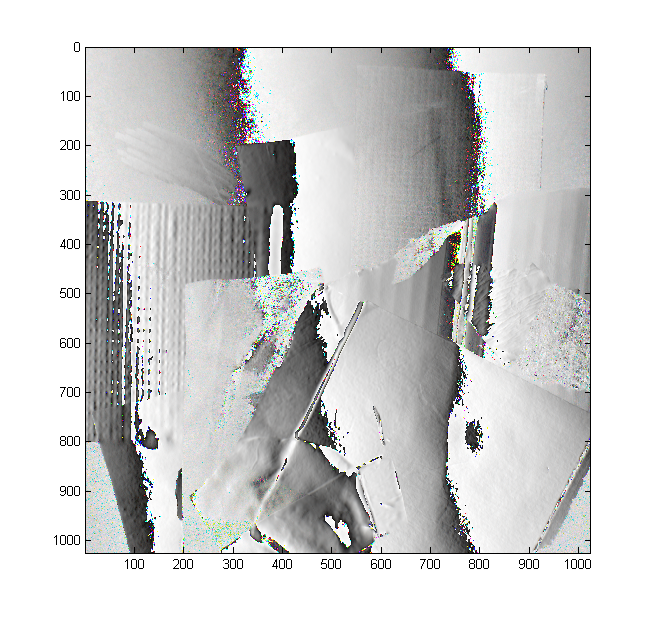
One could extract quite a bit of useful heuristic information out of this sort of near-raw data itself, if accuracy is not critical and the surfaces are glossy (it is the mixing of diffuse and specular that makes it difficult.) In fact, we base our initial guess on similar reasoning.